Goal
This project, involving researchers analysing data from Pfizer’s ATLAS database and supported by the Vivli AMR Surveillance Data Challenge and the ADILA project, aims to enhance real-time antimicrobial resistance (AMR) surveillance using flexible spatiotemporal modelling.
Lead
Kasim Allel Henriquez – Nuffield Department of Primary Care Health Science, University of Oxford
What we did
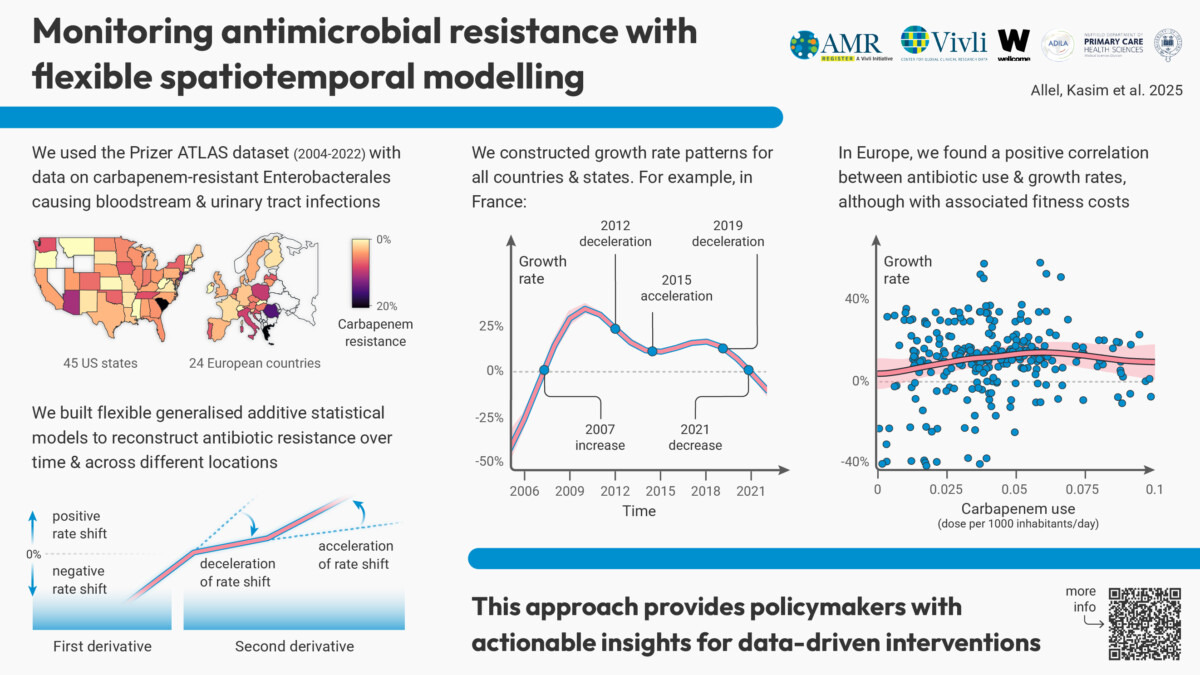
By examining 64,540 carbapenem-resistant Enterobacterales (CRE) isolates from bloodstream and urinary tract infections collected between 2004 and 2022, the study identifies changes in CRE growth rates and their association with time and geography in US-states and European countries. Utilising generalised additive models (GAMs) with Gaussian Markov Random Fields, the project provides policymakers with actionable insights for targeted interventions via identifying early change points in CRE growth rates. In subgroup analysis covering 24 European countries, the study also analyses CRE growth rates and carbapenem usage patterns to understand regional variations and inform an assessment of the relationship between these two variables.
Key learnings
Our study aims to optimise resource allocation and improve the effectiveness of AMR interventions by identifying early change points in resistance growth as potential early-warning signals.
Outputs
Funder
The Wellcome Trust and Vivli AMR Data Challenge